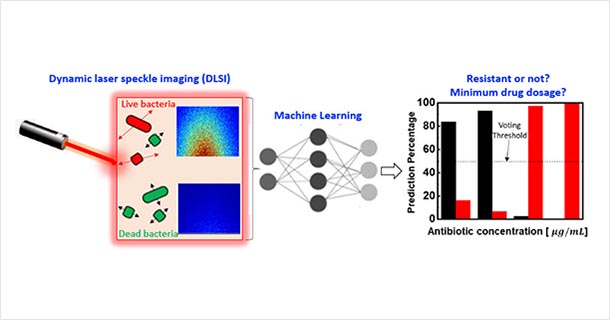
Convergence of machine learning and dynamic laser speckle imaging to monitor bacterial motion enables rapid and accurate identification of antibiotic resistance and the minimum drug dosage needed to inhibit bacterial growth. IMAGE: KEREN ZHOU, PENN STATE
Inexpensive and rapid testing of drugs for resistant infections possible
10/15/2020
By Walt Mills
UNIVERSITY PARK, Pa. — A rapid and simple method for testing the efficacy of antibacterial drugs on infectious microbes has been developed and validated by a team of Penn State researchers.
Antimicrobial resistant infection is one of the major threats to human health globally, causing 2.5 million infections and 35,000 deaths annually, with the potential to grow to 10 million deaths annually by 2050 without improved techniques for detection and treatment.
Several rapid testing techniques have been developed, but they do not live up to the reliability of the gold standard technology, which requires 18 to 24 hours for reliable results. In many cases, patients need to be treated with antibiotics in a crisis, leading clinicians to prescribe broad-spectrum antibiotics that may actually lead to greater drug resistance or unacceptable side effects.
“Compared to other methods of detection, our method does not require complex systems and measurement setups,” says Aida Ebrahimi, assistant professor of electrical engineering and a senior author on a paper recently posted online in the journal ACS Sensors. “Its simplicity and low cost are among the advantages and coupling our technology to machine learning makes the accuracy of our method comparable to the gold standard method and much better than other rapid methods.”
The team tested their method against three strains of bacteria, including a resistant strain, to prove its effectiveness in the lab. Upon further development and validation with a broader range of pathogens and antibiotics, their method can allow physicians to prescribe the minimum dosage of the necessary drug, called the minimum inhibitory concentration (MIC) in a timely fashion.
A phenomenon that other tests fail to account for is that bacteria may initially appear to be dead, but then can revive and multiply after many hours. The team’s technology, augmented by machine learning, can predict whether the bacteria will revive or are actually dead, which is critical for accurate determination of the MIC value.
Their technique is called dynamic laser speckle imaging.
“The main advantages of our method are the speed and simplicity," explained Zhiwen Liu, professor of electrical engineering and the second corresponding author. You shine a laser beam on the sample and get all of these light scattering speckles. We can then capture these images and subject them to machine learning analysis. We capture a series of images over time, which is the dynamic part. If the bacteria are alive, you are going to get some motion, such as a small vibration or a little movement. You can get reliable, predictive results quickly, for example within one hour.”
In addition to the immediate benefits provided to the patient, the lower concentration of drugs entering the water supply translates to less pollution to the environment, he says.
“One of the exciting aspects of this research has been its multidisciplinary nature. As an electrical engineer, I find it quite fascinating to work on designing and developing an optical diagnostic system as well as performing microbiology assays,” said Keren Zhou, the co-lead first author on the paper and a doctoral student in electrical engineering.
His co-lead author, doctoral student Chen Zhou, added, “We plan to further develop our technique to a low-cost and portable platform, which would be especially beneficial for resource-limited settings.”
In this work, the researchers also collaborated with Jasna Kovac, assistant professor of food science, to validate their findings with gold standard microbiology methods.
Additional authors on the paper, titled “Dynamic Laser Speckle Imaging meets Machine Learning to enable Rapid Antibacterial Susceptibility Testing (DyRAST),” are Anjali Sapre, Jared Henry Pavlock, Ashley Weaver, Ritvik Muralidharan, Joshua Noble and Taejung Chung, all of Penn State.
Support for this work was provided by the Materials-Life Science Convergence Award sponsored by the Materials Research Institute, Huck Institutes of Life Sciences, College of Engineering and College of Medicine (Hershey) at Penn State. Other agencies providing partial support of the personnel include the National Science Foundation and the United States Department of Agriculture.
Penn State has applied for a patent on this technology.



